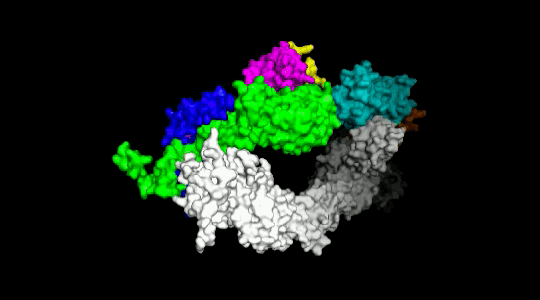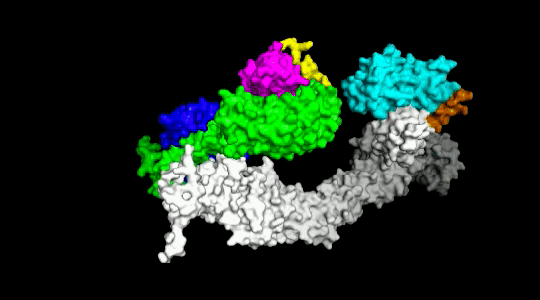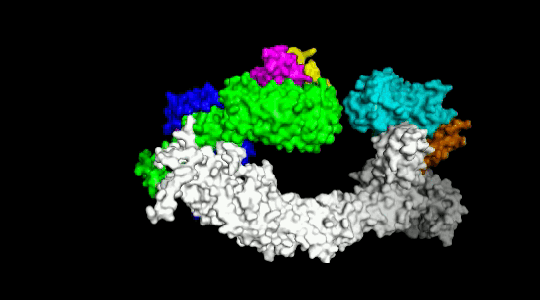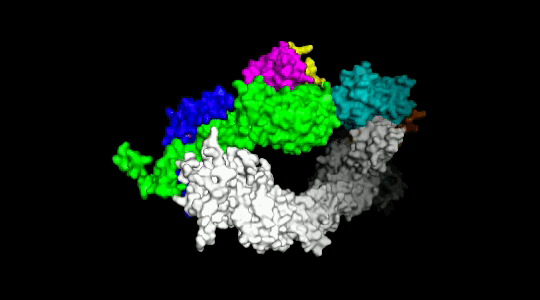
Over the years we have developed a variety of novel computational strategies such as ENTess, PolyPharma, EnsembleGen, miRmo, and HIVP-SAE, by integrating cheminformatics, bioinformatics, and systems chemical biology approaches. More recently as witnessed in the resurgence of artificial intelligence (AI), we are developing an utterly novel image-based AI platform (termed AI4MI) to predict direct molecular interactions (protein-protein, protein-RNA/DNA, and protein-small interactions) which are key to understanding cell signaling, disease progression, and therapeutic treatment. Such technologies are particularly invaluable when applied to an emerging field, for instance, protein-miRNA interactions, where little information is available. Along the same line, we combined the large amount of cell line and patient genomic profiles and chemical compound structure-activity data to build AI models (termed DL4DR) that enables us to deep screen for drug responses in all cancer cell lines (>1,500) and ultimately translate to personalized patient treatment. We expect that these technologies are game changers by minimizing, or even eliminating, the need of costly and time-consuming laboratory tests for molecular interactions or drug responses. They will also provide more complete pictures of molecular signaling and disease-driving mechanisms.
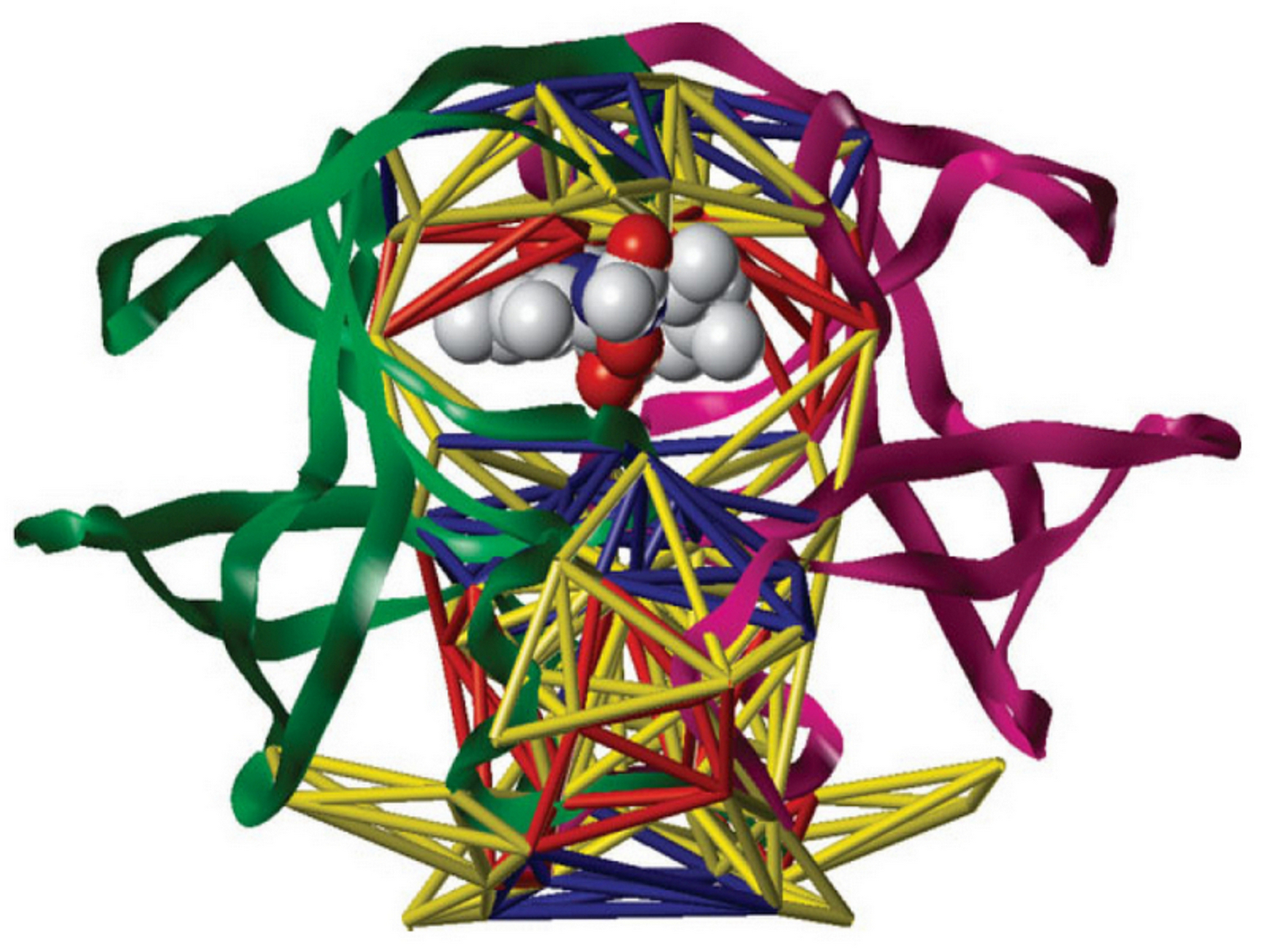
We are especially interested in targeting those traditionally viewed undruggable molecular pathways. For instance, it has been revealed that S-phase kinase-associated protein 2 (Skp2) is overexpressed in many cancers and represents a molecular marker for poor prognosis. Moreover, Skp2 is a known E3 ligase of a tumor suppressor p27, but it can also trigger non-proteolytic K63-linked ubiquitination of oncogenic Akt, which facilitates Akt activation, leading to increased cancer glycolysis. In addition, Skp2 inactivation induces a massive cellular senescence and/or apoptosis response in a p19Arf/p53-independent but p27-dependent manner. Therefore, we developed first-in-class small molecule inhibitors which exhibit potent anti-tumor activities through specific Skp2 targeting (Cell. 2013) as a promising "pro-senescence/apoptosis" and "anti-glycolysis" approach to cancer therapy. Currently we are conducting IND-enabling studies for regulatory filing and first-in-human trials. Our other high impact publications include (Nat. Commun 2022; JACS 2021; Nature 2019; Nat Commun 2018; Nat Cell Biol. 2017; Nat Commun 2017; etc.).
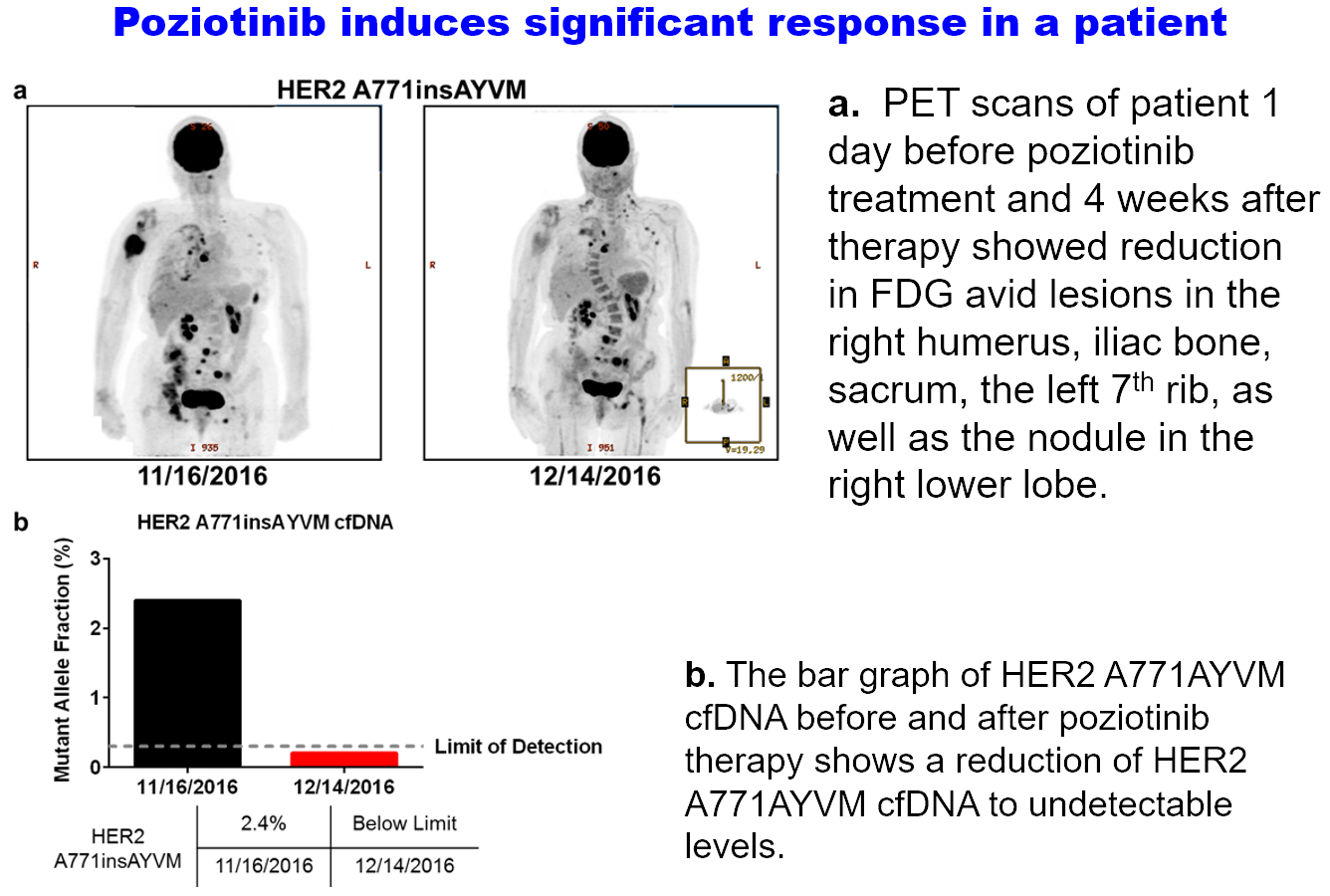
With our multi-modal approaches based on the latest AI technologies and chemo-genomic approaches, we have been very fruitful in polypharmacology studies and drug repurposing. One of the most success stories is the discovery of a previously shelfed drug, poziotinib, as a potent and selective inhibitor of EGFR/HER2 exon-20 insertion mutants. In a further Phase 2 clinical trial, the drug shrunk tumors significantly in eight of 11 NSCLC patients (Nat. Med. 2018). With such exciting result, MD Anderson and Spectrum Pharmaceuticals signed a licensing agreement, and the study is being continued as one of the largest Phase II clinical trials in the world for this subset of NSCLC patient population.
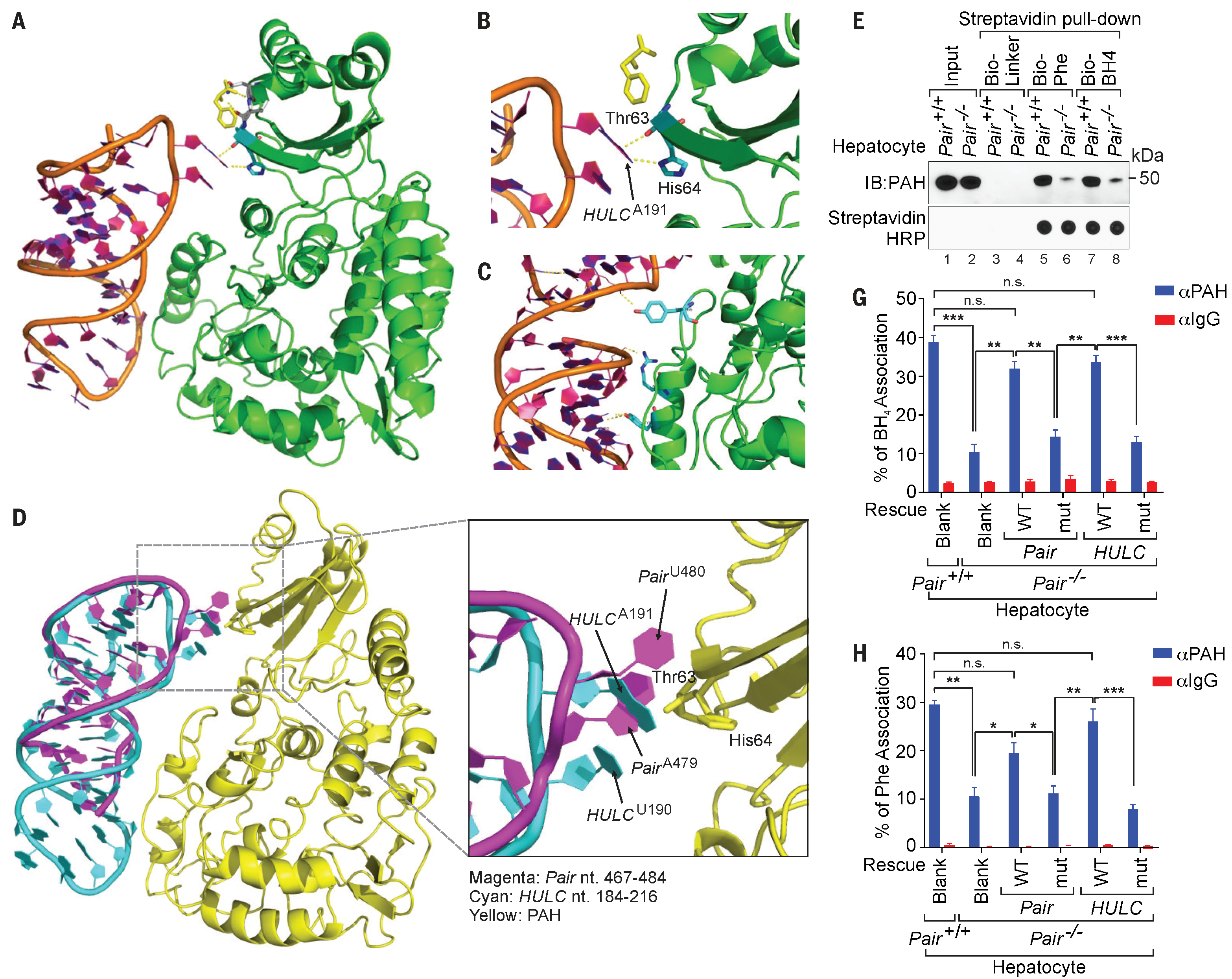
In addition to cancer, we are interested in other diseases such as metabolic disorders and COVID-19. For instance, phenylketonuria (PKU) is a single-gene metabolic disorder that needs to be screened in all newborns. However, currently the therapeutic option to treat this disease is very limited, and its neurological toxic effects can only be prevented by dietary supplement. Recently we identified a lncRNA in human, termed HULC, that interacts with phenylalanine hydroxylase (PAH) and modulates its functions. Using our commputational platform, we elucidated how HULC structurally synergizes with a regulatory phenylalanine to enhance metabolism of the substrate phenylalanine. Such novel insight will help modify lncRNAs to treat phenylketonuria. This exciting discovery was published on Science. 2021.