miRmo - A web server for microRNA motif screening
miRmo is a computational tool that enables fast motif screening in pre-microRNA (pre-miRNA) secondary structures. This may help conduct function annotation of miRNAs and elucidate the potential off-targeting properties of therapeutic agents that target those non-coding molecules.
miRmo Input
miRmo Output
This is where the link to the results will show up after the screening job is submitted.
Frequently Asked Questions (FAQ)
MicroRNAs (miRNAs) are a class of short (18-24 nt), non-coding RNAs that are widely conserved among many known species.
These molecules are not translated into proteins (thus non-coding) but they can mediate post-transcriptional gene expression regulation by targeting mRNAs or interacting with other molecules. Via these interactions, miRNA play various important roles in living organisms.
miRmo is a user-friendly miRNA motif screening tool, which is implemented as an easy-to-access web server. The program
is developed primarily using Python and PHP. Users can input the target motifs by following certain rules. Meanwhile, the motif types and databases to query can also be specified.
miRmo can screen RNA databases to identify all all RNAs that contain a specified non-paired secondary structure motif. This program quickly searches the predicted 2D structures of pre-miRNAs for a given secondary structural motif and then provides a list of miRNAs, the locations of the motif in the pre-miRNAs, CT files of the predicted pre-miRNA structures, and 2D images of the structures.
This serves two purposes: 1). A known motif is important in one RNA, but investigators want to further understand whether this motif exists in other RNAs (thus possessing other functions). 2). The motif is well known and has been targeted for therapeutics development. The investigators would like to know whether such targeting is specific; if not, what are the potential off-targeting RNAs and what are the possible side effects?
This serves two purposes: 1). A known motif is important in one RNA, but investigators want to further understand whether this motif exists in other RNAs (thus possessing other functions). 2). The motif is well known and has been targeted for therapeutics development. The investigators would like to know whether such targeting is specific; if not, what are the potential off-targeting RNAs and what are the possible side effects?
miRmo is unique and different from most of other RNA motif search programs in several aspects:
- It is NOT designed to search for NEW or UNKNOWN motifs; rather, users need to have some initial understanding of the motifs (e.g., pairing patterns, some biological functions, etc.) and would like to know more about the functional implications of the motif in many other RNAs.
- It is NOT to screen matured miRNAs; instead, it is implemented to screen precursors which have more structural features for their functions.
- It does NOT need miRNA 3D structures. Only the primary sequence and secondary structure information is used. However, the 3D information, if available, can help interpret the functions of miRNAs. This is why we provide PDB Search capability as described below.
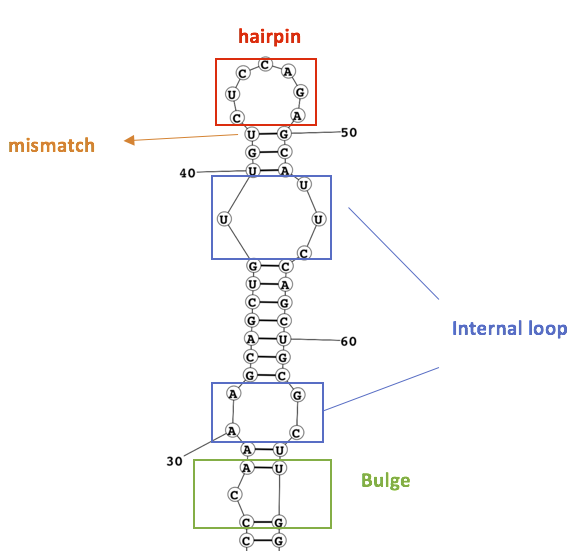
Currently, miRmo can search for Internal Loop, Bulge, Mismatch, and Hairpin motifs.
Knowing the 3D structures of RNAs is crucial to elucidate their functions. However, most pre-miRNAs do not have solved 3D structures. The PDB Search option enables users to identify all RNAs with the determined 3D structures containing the queried motif, and this may provide some hints on the 3D structure of the queried motif and thus its functions in pri-miRNAs. However, please be advised that hints are just hints; they might not be conclusive or sometimes even misleading. Thus, investigators still need their expert knowledge to interpret the data.
Anyone - miRmo is a user-friendly and open-access service. Currently no registration is needed to explore its full functionality. However, commercial users should contact the author for agreement and support.
If you used miRmo in your work, please cite our web server (https://www.imdlab.net/mirmo/mirmo.php), or our manuscript once it is published.
Please contact us at shuxing[at]molsoftz[dot]com.
